Professional Documents
Culture Documents
Cervigram Image Segmentation Based On Reconstructive Sparse Representations
Uploaded by
Prasanalakshmi BalajiOriginal Title
Copyright
Available Formats
Share this document
Did you find this document useful?
Is this content inappropriate?
Report this DocumentCopyright:
Available Formats
Cervigram Image Segmentation Based On Reconstructive Sparse Representations
Uploaded by
Prasanalakshmi BalajiCopyright:
Available Formats
Cervigram Image Segmentation Based On Reconstructive Sparse Representations
Shaoting Zhang1 , Junzhou Huang1 , Wei Wang2 , Xiaolei Huang2 , Dimitris Metaxas1 Rutgers University 110 Frelinghuysen Road Piscataway, NJ 08854, USA 2 Department of Computer Science and Engineering Lehigh University, Bethlehem, PA 18015, USA
ABSTRACT
We proposed an approach based on reconstructive sparse representations to segment tissues in optical images of the uterine cervix. Because of large variations in image appearance caused by the changing of the illumination and specular reection, the color and texture features in optical images often overlap with each other and are not linearly separable. By leveraging sparse representations the data can be transformed to higher dimensions with sparse constraints and become more separated. K-SVD algorithm is employed to nd sparse representations and corresponding dictionaries. The data can be reconstructed from its sparse representations and positive and/or negative dictionaries. Classication can be achieved based on comparing the reconstructive errors. In the experiments we applied our method to automatically segment the biomarker AcetoWhite (AW) regions in an archive of 60,000 images of the uterine cervix. Compared with other general methods, our approach showed lower space and time complexity and higher sensitivity. Keywords: segmentation, cervix image, biomarker AcetoWhite, reconstructive errors, sparse representation, K-SVD, OMP
1 CBIM,
1. INTRODUCTION
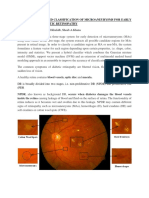
Segmentation of dierent regions of medical images can assist doctors to analyze them. Area information from segmentation is important in many clinical cases. In this work, we propose an approach to automatically segment the biomarker AcetoWhite (AW) regions in an archive of 60,000 images of the uterine cervix. These images are optical cervigram images acquired by Cervicography using specially-designed cameras for visual screening of the cervix (Figure 1). They were collected from the NCI Guanacaste project1 for the study of visual features correlated to the development of precancerous lesions. The most important observation in a cervigram image is the AW region, which is caused by whitening of potentially malignant regions of the cervix epitheliuem, following application of acetic acid to the cervix surface. Since the texture, size and location of AW regions have been shown to correlate with the pathologic grade of disease severity, accurate identication and segmentation of AW regions in cervigrams have signicant implications for diagnosis and grading of cervical lesions. However, accurate tissue segmentation in cervigrams is a challenging problem because of large variations in the image appearance caused by the changing of illumination and specular reection in pathology. As a result, the color and texture features in optical images often overlap with each other and are not linearly separable when training samples are larger than a certain level (Figure 2).
1.1 Related work
Previous work on cervigram segmentation has reported limited success using K-means clustering,2 Gaussian Mixture Models,3 Support Vector Machine (SVM) classiers.4 Shape priors are also proposed.5 Although such priors are applicable to cervix boundary, it does not work well with AW since AW regions may have arbitrary shapes. Supervised learning based segmentation,67 holds promise, especially with increasing number of features. However, because of the intrinsic diversity between images and the overlap between feature distributions of
{shaoting, jzhuang, dnm}@cs.rutgers.edu, {wew305, xih206}@lehigh.edu
Figure 1. Examples of digitized cervicographic images (i.e. cervigrams) of the uterine cervix, created by the National Library of Medicine (NLM) and the National Cancer Institute (NCI). Ground truth boundary markings by 20 medical experts. Our work is aimed for automatically segmenting the biomarker AcetoWhite (AW) regions, which indicates clinical signicance.
Figure 2. Color distribution of AW and non-AW regions. Red means AW and blue means non-AW. The left one comes from one sample. The right one comes from one hundred samples.
dierent classes, it is dicult to learn a single classier that can perform tissue classication with low error rate for a large image set. Our empirical evaluation shows that overtting is a serious problem when training a single (SVM) classier using all training samples the average sensitivity of the classier is relatively low. A potential solution is to use a Multiple Classier System (MCS),8 which trains a set of diverse classiers that disagree on their predictions and eectively combines the predictions in order to reduce classication error. Voting, AdaBoost, bagging and STAPLE9 can be employed. A necessary condition for the above ensemble methods is that all the base classiers should provide suciently good performance, usually 50% or higher sensitivity and specicity in order to support the ensemble. However, there may be large variance in base classier performance in our case. Some classiers commonly have lower sensitivity than 50%. Wang and Huang10 proposed a method to nd the best base classier based on distance guided selection, which achieves state-of-the-art results in a subset of the archive. In our method we focus on nding a single classier by transforming the data to a higher dimension with sparse constraints. Then the data can be more separated using sparse representations. This classier is potentially useful for MCS since the sensitivity and specicity are always larger than 50% in our experiments. Finding the sparse representation typically consists of the sparse coding and codebook update. Greedy algorithms such as matching pursuit (MP)11 and orthogonal matching pursuit (OMP)12 can be employed for nding sparse coecients (coding). Extensive study of these algorithms shows that if the sought solution is sparse enough, these greedy techniques can obtain the optimal solution.13 When the sparse data has group clustering trend,14 AdaDGS15 can be employed to further improve the performance. To update codebook, method of optimal direction (MOD)16 and K-SVD17 are two eective approaches. Although both of them result in similar results, we prefer K-SVD because of its better convergence rate. After nding positive and negative dictionaries from training images and computing sparse coecients from testing images, reconstructive errors can be obtained and compared. The pixel can be assigned to the class with lower errors. Our main contributions are the following: 1) we introduce the reconstructive sparse representations and K-SVD algorithms to the medical imaging community, which are originated from the compressive sensing eld; 2) we apply this theory to solve the challenging cervigram image segmentation problem and achieve improved performance. Details of this method is explained in Section
Figure 3. The algorithm framework. The left column represents the data. The right column represents algorithms including K-SVD.
2 and experiments are shown in Section 3.
2. METHODOLOGY 2.1 Framework
Figure 3 illustrates the algorithm framework. In the training stage, ground truth is manually obtained by clinical experts. Patches on the ground truth are labeled as positive ones, while the others are negative ones. These patches are fed into K-SVD to generate positive and negative dictionaries. Then using these two dictionaries, the sparse coding step is applied on patches extracted from testing images to compute two sets of sparse coecients. From the coecients and corresponding dictionaries, reconstructive errors are calculated and compared for classication. An alternative way is to classify the sparse coecients (using SVM) instead of classifying the original data, which is also tested in Section 3. Details of the reconstructive sparse representation are discussed in Section 2.2 and Section 2.3.
2.2 Learning sparse dictionaries
Reconstructive sparse representation is used here to classify image patches. The objective of sparse representation is to nd D and X by minimizing the following equation:
min{ Y DX
D,X
2 F}
subject to i, xi
(1)
Where Y represents signals (image patches here), D is the overcomplete dictionary, X is the sparse coecients, 0 is the l0 norm counting the nonzero entries of a vector, F is the Frobenius norm. Denote yi as the ith column of Y , xi as the ith column of X , then yi and xi are the ith signal vector and coecient vector respectively, with dimensionality D Rnk , yi Rn and xi Rk . K-SVD algorithm starts from a random D and X obtained from the sparse coding stage. The sparse coding stage is based on pursuit algorithms to nd the sparse coecient xi for each signal yi . OMP is employed in this stage. OMP is an iterative greedy algorithm that selects at each step the dictionary element that best correlates with the residual part of the signal. Then it produces a new approximation by projecting the signal onto those elements already selected.13 The algorithm framework of OMP is listed in the Table 1.
Table 1. The framework of the OMP algorithm.
Input: dictionary D Rnk , input data yi Rn Output: coecients xi Rk = Loop: iter=1, ..., L Select the atom which most reduces the objective arg min
j
min |yi D{j } x |2 2
x
(2)
Update the active set: {j } Update the residual using orthogonal projection
T T r I D (D D )1 D yi
(3)
Update the coecients
T T x = (D D )1 D yi
(4)
In the codebook update stage K-SVD employs a similar approach as K-Means to update D and X iteratively. In each iteration D and X are xed except only one column di and the coecients corresponding to di (ith row in X ), denoted as xi T . The Equation 1 can be rewritten as
2 2
= Y
j =i
dj xj T
2 F
Y
j =1
dj xj T
F
di xi T
F
(5) (6)
= Ei di xi T
i We need to minimize the dierence between Ei and di xi T with xed Ei , by nding alternative di and xT . Since SVD nds the closest rank-1 matrix that approximates Ei , it can be used to minimize the Equation 5. Assume Ei = U V T , di is updated as the rst column of U , which is the eigenvector corresponding to the largest eigenvalue. xi T is updated as the rst column of V multiplied by (1, 1).
However, the updated xi T may not be sparse anymore. The solution is logical and easy. We just discard the zero entries corresponding to the old xi T . The detail algorithms of K-SVD are listed in the table 2.
2.3 Reconstructive errors
Using K-SVD algorithm we can obtain two dictionaries for positive patches and negative patches separately, denoted as D+ and D respectively. The simplest strategy to use dictionaries for discrimination is to compare the errors of a new patch y reconstructed by D+ and D and choose the smaller one as its type, as shown in equation 9.
type = arg min{ y Di x 2 2 } subject to x
i=+,
(9)
The potential problem of this method is that the dictionaries are trained separately. That is to say, positive/negative dictionary only depends on positive/negative patches, so it attempts to reconstruct better for
Table 2. The framework of the K-SVD algorithm.
Input: dictionary D Rnk , input data yi Rn and coecients xi Rk Output: D and X Loop: Repeat until convergence Sparse coding: use OMP to compute coecient xi for each signal yi , to minimize min{ yi Dxi 2 2 } subject to xi
xi 0
(7)
Codebook update: for i = 1, 2, ..., k , update each column di in D and also xi T (ith row) Find the group using di (xi T = 0), denoted as i Compute error matrix Ei as in equation 5 Restrict Ei by choosing columns corresponding to i . The resized error is R denoted as Ei Apply SVD and obtain
R Ei = U V T
(8)
Update di as the rst column of U . Update nonzero elements in xi T as the rst column of V multiplied by (1, 1)
positive/negative patches but not worse for negative/positive patches. Discriminative methods can be considered to alleviate this problem.18 However, the tuning parameters of the discriminative system are very sensitive and it can only converge within small intervals. In our case, reconstructive method works relatively well. Discriminative method with this application is left to future investigation. An alternative way is to classify the sparse coecients x instead of y . x from training images can be fed into SVM or other classier. The intuition is that x is in higher dimension with sparse constants and can be more separated. Both of these two approaches are tested in Section 3.
2.4 Tracing regions
Since there is no shape information considered, the resulting areas are usually disconnected. Inspired by the edge linking stage of Canny edge detector, similar procedure can also be applied on this application. Equation 9 can be rewritten as:
error = y D x
2 2
y D+ x
2 2
(10)
When error < 0, the testing data is assigned to the negative samples. Otherwise it is positive. However, due to noise, there may be positive instances below the threshold (0). Thus similar to the Canny edge detector, two thresholds T1 and T2 (T1 > T2 ) can be predened. In the rst pass, T1 = 0 is used as the threshold and classication is performed. This procedure is the same as Section 2.2. In the second pass, T2 < 0 is set as the new threshold. The errors of neighboring points of the rst results are checked, and the points with error > T2 are merged into the positive samples. With ideal thresholds, the disconnectivity problem can be alleviated in a certain level. However, the value of T2 highly depends on the application and currently is found by cross validation and brute force. Starting from 0, T2 is decreased by a small step each time. The sensitivity and specicity are computed in each step. The parameters causing the best performance are chosen. More sophisticated approaches are left for future investigations.
(a) AW dictionaries
(b) non-AW dictionaries
Figure 4. positive (AW) and negative (non-AW) dictionaries trained from K-SVD. Each dictionary is displayed as 16 by 16 cells (256 cells). Each cell comes from the column of the dictionary and is reshaped as a 5 by 5 patch. Some patches from dierent dictionaries are quite similar. Note that these patches do not directly represent image patches, since the columns are normalized in HSV color space.
Table 3. Performance comparison between 4 classiers, measured by the mean of sensitivity and specicity.
Sensitivity SVM with image patches Nearest Neighbor Compare reconstructive errors SVM with sparse coecients 50.24% 55.62% 62.71% 61.46%
Specicity 63.59% 69.18% 75.85% 76.37%
2.5 Implementation details
Cervigram images from the NCI/NLM archive with multiple-expert boundary markings are available for training and validation purposes. 100 images of diverse appearance were selected for training and testing. To maximally mix the samples, 10 image is used for testing and validation and the remaining 90 ones are used for training. The mean sensitivity and specicity are reported. Dierent color spaces including RGB, HSV and Lab are tested. HSV is chosen since it is slightly better. Other color spaces, texture and appearance information can also be considered. Each patch is a 5 by 5 square centered in the pixel and concatenated H,S,V information into single vectors (75 by 1, n = 75). We choose the sparse factor L = 6 and dictionaries of size k = 256. Although there are many choices for these values, they are not arbitrary. They need to satisfy the constraints mentioned in19 to guarantee convergence. In each image 1,000 patches are randomly selected from both AW and non-AW regions, 500 for each. Overall 90,000 patches are generated from the training images. 60 iterations of K-SVD are performed. The positive and negative dictionaries represent AW and non-AW regions respectively. Each column of dictionaries are reshaped as 5 by 5 patches, and they are displayed together in Figure 4. Some patches from dierent dictionaries are similar, proving that classication is a challenging task. Utilizing sparse representations can alleviate this problem.
3. RESULTS
The method was implemented in Matlab R2009a and tested on a 2.40 GHz Intel Core2 Quad computer with 8G RAM. It was compared with SVM and k nearest neighbors. SVM failed to handle 90,000 patches since it would consume most memories and couldnt converge. Thus the data for SVM was down sampled. Instead of feeding image patches into SVM, we also trained SVM using sparse coecients. Nearest neighbor method was also time and space consuming because of the large training set. K-SVD was more ecient with 5 seconds for each iteration and less than 1GB RAM because of its sparsity. Table 3 shows the results of dierent classiers measured by sensitivity and specicity. Figure 5 visualizes the segmentation results of a specic image. Since the distributions of image patches were overlapped (Figure 2),
Figure 5. Results of a specic image. From left to right: original image; ground truth; SVM with image patches; nearest neighbor; reconstructive errors (equation 9).
SVM with image patches underperformed the classication task. Using sparse coecients in higher dimension, SVM performed better since the data was more separated. Although comparing reconstructive errors also achieved good results, it still had many noises from non-AW regions. The reason is that the K-SVD algorithm is not discriminative and there is no shape information considered. From the comparisons in Table 3 and Figure 5 we can nd that sparse representations are very useful in this challenging problem.
4. CONCLUSIONS
In this paper we proposed classiers based on reconstructive sparse representations to segment tissues in optical images of the uterine cervix. Our method was compared with some typical learning methods and worked better in this challenging case. In the future we would like to put eorts on three directions. 1) We will develop discriminative sparse representations instead of reconstructive ones. The dictionaries should perform better for its own class and also worse for other classes. Current discriminative methods highly depend on the parameters and are not stable. It can be further improved. 2) We will add our method into best base classier10 to improve the sensitivity and specicity of the whole system. 3) Combining sparse representations with SVM is also interesting.
5. ACKNOWLEDGEMENTS
The authors would like to thank the Communications Engineering Branch, National Library of Medicine-NIH, and the Hormonal and Reproductive Epidemiology Branch, National Cancer Institute-NIH, for providing the data and support of this work. The authors are thankful to Hongsheng Li (Lehigh), Tian Shen (Lehigh), Xiaoxu Wang (Rutgers), Ming Jin (Rutgers), Yuchi Huang (Rutgers) and Yang Yu (Rutgers) for stimulating discussions.
REFERENCES
[1] Jeronimo, J., Long, L., Neve, L., Bopf, M., Antani, S., and Schiman, M., Digital tools for collecting data from cervigrams for research and training in colposcopy, in [Colposcopy, J Low Gen Tract Disease ], 1625 (2006). [2] Tulpule, B., Hernes, D., Srinivasan, Y., Yang, S., Mitra, S., Sriraja, Y., Nutter, B., Phillips, B., Long, L., and Ferris, D., A probabilistic approach to segmentation and classication of neoplasia in uterine cervix images using color and geometric features, in [SPIE, Medical Imaging: Image Processing ], 9951003 (2005). [3] Gordon, S., Zimmerman, G., Long, R., Antani, S., Jeronimo, J., and Greenspan, H., Content analysis of uterine cervix images: Initial steps towards content based indexing and retrieval of cervigrams, in [SPIE, Medical Imaging: Image Processing ], 20372045 (2006). [4] Huang, X., Wang, W., Xue, Z., Antani, S., Long, L. R., and Jeronimo, J., Tissue classication using cluster features for lesion detection in digital cervigrams, in [SPIE, Medical Imaging: Image Processing ], (2008).
[5] Gordon, S., Lotenberg, S., and Greenspan, H., Shape priors for segmentation of the cervix region within uterine cervix images, in [SPIE, Medical Imaging: Image Processing ], (2008). [6] Shotton, J., J.Winn, Rother, C., and Criminisi, A., Joint appearance, shape and context modeling for multi-class object recognition and segmentation, in [ECCV ], (2006). [7] Schro, F., Criminisi, A., and Zisserman, A., Singlehistogram class models for image segmentation, in [ICVGIP ], (2006). [8] Artan, Y. and Huang, X., Combining multiple 2v-svm classiers for tissue segmentation, in [ISBI ], 488 491 (2008). [9] Wareld, S., Zou, K., and Wells, W., Simultaneous truth and performance level estimation (staple): An algorithmfor the validation of image segmentation, in [IEEE Trans. on Medical Imaging ], 903921 (2004). [10] Wang, W. and Huang, X., Distance guided selection of the best base classier in an ensemble with application to cervigram image segmentation, in [MMBIA ], (2009). [11] Mallat, S. and Zhang, Z., Matching pursuits with time-frequency dictionaries, in [IEEE Trans. Signal Process ], 33973415 (1993). [12] Chen, S., Billings, S. A., and Luo, W., Orthogonal least squares methods and their application to non-linear system identication, in [Intl. J. Contr ], 18731896 (1989). [13] Tropp, J. A., Greed is good: Algorithmic results for sparse approximation, in [IEEE Trans. Inf. Theory ], 22312242 (2004). [14] Huang, J. and Zhang, T., The benet of group sparsity, in [Technical Report arXiv:0901.2962, Rutgers University ], (2009). [15] Huang, J., Huang, X., and Metaxas, D., Learning with dynamic group sparsity, ICCV (2009). [16] Engan, K., Aase, S. O., and Hakon-Husoy, J. H., Method of optimal directions for frame design, in [IEEE Int. Conf. Acoust., Speech, Signal Process ], 24432446 (1999). [17] Aharon, M., Elad, M., , and Bruckstein, A., K-svd: An algorithm for designing overcomplete dictionaries for sparse representation, in [IEEE Trans. Signal Process ], 43114322 (2006). [18] Mairal, J., Bach, F., Ponce, J., Sapiro, G., and Zisserman, A., Discriminative learned dictionaries for local image analysis, in [CVPR ], 43114322 (2008). [19] Starck, J. L., Elad, M., and Donoho, D., Image decomposition via the combination of sparse representations and a variational approach, IEEE Trans. on Image Processing 14, 15701582 (2004).
You might also like
- Breast Cancer ClassificationDocument18 pagesBreast Cancer ClassificationSatwik Sridhar ReddyNo ratings yet
- Vishal M. Patel, Tao Wu, Soma Biswas, P. Jonathon Phillips and Rama ChellappaDocument4 pagesVishal M. Patel, Tao Wu, Soma Biswas, P. Jonathon Phillips and Rama ChellappaSatish NaiduNo ratings yet
- Segmentation of Brain MR Images Based On Finite Skew Gaussian Mixture Model With Fuzzy C-Means Clustering and EM AlgorithmDocument9 pagesSegmentation of Brain MR Images Based On Finite Skew Gaussian Mixture Model With Fuzzy C-Means Clustering and EM AlgorithmNuthangi MaheshNo ratings yet
- Biomedical Data Analysis by Supervised Manifold LearningDocument4 pagesBiomedical Data Analysis by Supervised Manifold LearningCarlos López NoriegaNo ratings yet
- Asilomar Bhalerao Anand SaravananDocument3 pagesAsilomar Bhalerao Anand SaravanansosoNo ratings yet
- MIA PresentationDocument8 pagesMIA Presentationahsan puriNo ratings yet
- Isbi 2016 7493382Document4 pagesIsbi 2016 7493382Gautham SgNo ratings yet
- Sujeet Bhagwat (04) - Aditya Gujar (27) - Swaraj Govindwar (26) - Krishna GatlewarDocument25 pagesSujeet Bhagwat (04) - Aditya Gujar (27) - Swaraj Govindwar (26) - Krishna GatlewarSwaraj GovindwarNo ratings yet
- Osteoporosis Detection Using Machine and Deep Learning TechniquesDocument15 pagesOsteoporosis Detection Using Machine and Deep Learning TechniquesSeddik KhamousNo ratings yet
- Samulski Classification07Document11 pagesSamulski Classification07Perry GrootNo ratings yet
- A Survey of Image Processing Techniques For Detection of MassDocument6 pagesA Survey of Image Processing Techniques For Detection of MassInternational Journal of Application or Innovation in Engineering & ManagementNo ratings yet
- An Intelligent Black Widow Optimization On Image Enhancement With Deep Learning Based Ovarian Tumor Diagnosis ModelDocument16 pagesAn Intelligent Black Widow Optimization On Image Enhancement With Deep Learning Based Ovarian Tumor Diagnosis ModelAgnesNo ratings yet
- Comparing Bayesian Models For Organ Contouring inDocument4 pagesComparing Bayesian Models For Organ Contouring inParakhModyNo ratings yet
- 25309-Article Text-29372-1-2-20230626Document8 pages25309-Article Text-29372-1-2-20230626Munib MesinovicNo ratings yet
- Dose-Reduction Experiment During A Cervical Vertebrae ExaminationDocument12 pagesDose-Reduction Experiment During A Cervical Vertebrae ExaminationDesty ArianiNo ratings yet
- CS 231N Final Project Report: Cervical Cancer ScreeningDocument9 pagesCS 231N Final Project Report: Cervical Cancer Screeningshital shermaleNo ratings yet
- Project 3: Technical University of DenmarkDocument10 pagesProject 3: Technical University of DenmarkRiyaz AlamNo ratings yet
- Brain Tumor Detection Using Machine LearningDocument9 pagesBrain Tumor Detection Using Machine LearningRayban PolarNo ratings yet
- Contour Detection and Hierarchical Image SegmentationDocument19 pagesContour Detection and Hierarchical Image SegmentationQuynhtrang NguyenNo ratings yet
- Mammogram Classification Using Law's Texture Energy Measure and Neural NetworksDocument6 pagesMammogram Classification Using Law's Texture Energy Measure and Neural NetworksMRFNo ratings yet
- Oversample Minority Classes in Lung Ultrasound Using Generative Adversarial NetworkDocument3 pagesOversample Minority Classes in Lung Ultrasound Using Generative Adversarial NetworkwangcosimoNo ratings yet
- Temporal Analysis of Mammograms Based On Graph Matching: 1 BackgroundDocument8 pagesTemporal Analysis of Mammograms Based On Graph Matching: 1 BackgroundAnand BalajiNo ratings yet
- Adaptive Segmentation Algorithm Based On Level Set Model in Medical ImagingDocument9 pagesAdaptive Segmentation Algorithm Based On Level Set Model in Medical ImagingTELKOMNIKANo ratings yet
- ReportDocument10 pagesReportOumaima El GhafikiNo ratings yet
- Application of PLS-DA in Multivariate Image AnalysisDocument9 pagesApplication of PLS-DA in Multivariate Image AnalysisIvan TrifkovićNo ratings yet
- Automatic Detection and Segmentation of Cortical Lesions in Multiple SclerosisDocument1 pageAutomatic Detection and Segmentation of Cortical Lesions in Multiple SclerosisSemana da Escola de Engenharia da Universidade do MinhoNo ratings yet
- 6614 Ijcsit 12Document8 pages6614 Ijcsit 12Anonymous Gl4IRRjzNNo ratings yet
- This Work Is Supported in Part by The Sensip Center and The Consortium Award No. S0322Document5 pagesThis Work Is Supported in Part by The Sensip Center and The Consortium Award No. S0322Prabhakar EleNo ratings yet
- Visualizing Conformal PredictionsDocument14 pagesVisualizing Conformal PredictionsAGNo ratings yet
- Karin Iccsa2012 v1Document33 pagesKarin Iccsa2012 v1Karin KomatiNo ratings yet
- Folmsbee Et Al. - 2018 - Active Deep Learning Improved Training Efficiency of Convolutional Neural Networks For Tissue Classification inDocument4 pagesFolmsbee Et Al. - 2018 - Active Deep Learning Improved Training Efficiency of Convolutional Neural Networks For Tissue Classification inCarlos Henrique LemosNo ratings yet
- 2005 - Designing Multiple Classi Er SystemsDocument10 pages2005 - Designing Multiple Classi Er SystemsluongxuandanNo ratings yet
- Linear Discriminant Analysis and Support Vector Machines For Classifying Breast CancerDocument4 pagesLinear Discriminant Analysis and Support Vector Machines For Classifying Breast CancerIAES IJAINo ratings yet
- Comparison of Algorithms For Segmentation of Blood Vessels in Fundus ImagesDocument5 pagesComparison of Algorithms For Segmentation of Blood Vessels in Fundus Imagesyt HehkkeNo ratings yet
- J. Appl. Math. & Informatics Vol. 26 (2008), No. 5 - 6, Pp. 861 - 876Document16 pagesJ. Appl. Math. & Informatics Vol. 26 (2008), No. 5 - 6, Pp. 861 - 876Sumanta DasNo ratings yet
- Obtaining Super-Resolution Images by Combining Low-Resolution Images With High-Frequency Information Derivedfrom Training ImagesDocument13 pagesObtaining Super-Resolution Images by Combining Low-Resolution Images With High-Frequency Information Derivedfrom Training ImagesAnonymous Gl4IRRjzNNo ratings yet
- Identifying Tuberculosis Type in CtsDocument8 pagesIdentifying Tuberculosis Type in Ctswork gradsNo ratings yet
- ScienceDocument7 pagesSciencePeter69No ratings yet
- Attention Based Graph ClassificationDocument5 pagesAttention Based Graph ClassificationAnđela TodorovićNo ratings yet
- Recognizing Imprecisely Localized, Partially Occluded and Expression Variant Faces From A Single Sample Per ClassDocument23 pagesRecognizing Imprecisely Localized, Partially Occluded and Expression Variant Faces From A Single Sample Per Classsimar rocksNo ratings yet
- School of Biomedical Engineering, Southern Medical University, Guangzhou 510515, ChinaDocument4 pagesSchool of Biomedical Engineering, Southern Medical University, Guangzhou 510515, ChinaTosh JainNo ratings yet
- Face Recog Using Wavelet Transformations PDFDocument5 pagesFace Recog Using Wavelet Transformations PDFRahul JainNo ratings yet
- Software Defect Identification Using MacDocument10 pagesSoftware Defect Identification Using Macwmarasigan2610No ratings yet
- SVM Quantile DNADocument9 pagesSVM Quantile DNAXiao YaoNo ratings yet
- Madooei2013 Chapter AutomaticDetectionOfBlue-WhiteDocument8 pagesMadooei2013 Chapter AutomaticDetectionOfBlue-WhitemaryNo ratings yet
- 5.1 Naive Bayes Experimental ResultDocument24 pages5.1 Naive Bayes Experimental ResultShafayet UddinNo ratings yet
- Statistical Learning Master ReportsDocument11 pagesStatistical Learning Master ReportsValentina Mendoza ZamoraNo ratings yet
- Gender FaceDocument9 pagesGender FacessanyiNo ratings yet
- Characterizing Architectural Distortion in Mammograms by Linear SaliencyDocument12 pagesCharacterizing Architectural Distortion in Mammograms by Linear SaliencySebastianNo ratings yet
- Machine Learning in MRDocument34 pagesMachine Learning in MR吉吉No ratings yet
- Electrical Engineering Technical Seminar ReportDocument19 pagesElectrical Engineering Technical Seminar Reportdastagir shahNo ratings yet
- Deep Lesion Tracker: Monitoring Lesions in 4D Longitudinal Imaging StudiesDocument11 pagesDeep Lesion Tracker: Monitoring Lesions in 4D Longitudinal Imaging StudiesMihir MehtaNo ratings yet
- Remote Sensing Image Classification ThesisDocument4 pagesRemote Sensing Image Classification ThesisPaperWritingServicesForCollegeStudentsOmaha100% (2)
- Comparison of Artificial Neural Network and Bayesian Belief Network in Computer-Assisted Diagnosis Scheme For MammographyDocument5 pagesComparison of Artificial Neural Network and Bayesian Belief Network in Computer-Assisted Diagnosis Scheme For MammographyUma GloriousNo ratings yet
- Paper 4Document14 pagesPaper 4Ramyaa PrasathNo ratings yet
- Research Reports Mdh/Ima No. 2007-4, Issn 1404-4978: Date Key Words and PhrasesDocument16 pagesResearch Reports Mdh/Ima No. 2007-4, Issn 1404-4978: Date Key Words and PhrasesEduardo Salgado EnríquezNo ratings yet
- Robust Fuzzy C-Mean Algorithm For Segmentation and Analysis of Cytological ImagesDocument3 pagesRobust Fuzzy C-Mean Algorithm For Segmentation and Analysis of Cytological ImagesWARSE JournalsNo ratings yet
- Random Sets Approach and Its ApplicationsDocument12 pagesRandom Sets Approach and Its ApplicationsThuy MyNo ratings yet
- A Method of Face Recognition Based On Fuzzy C-Means Clustering and Associated Sub-NnsDocument11 pagesA Method of Face Recognition Based On Fuzzy C-Means Clustering and Associated Sub-Nnsnadeemq_0786No ratings yet
- Project PlanDocument55 pagesProject PlanAlfredo SbNo ratings yet
- Group 7 Worksheet No. 1 2Document24 pagesGroup 7 Worksheet No. 1 2calliemozartNo ratings yet
- 1 Nitanshi Singh Full WorkDocument9 pages1 Nitanshi Singh Full WorkNitanshi SinghNo ratings yet
- DIAC Experienced Associate HealthcareDocument3 pagesDIAC Experienced Associate HealthcarecompangelNo ratings yet
- 2023 VGP Checklist Rev 0 - 23 - 1 - 2023 - 9 - 36 - 20Document10 pages2023 VGP Checklist Rev 0 - 23 - 1 - 2023 - 9 - 36 - 20mgalphamrn100% (1)
- Mental Status ExaminationDocument34 pagesMental Status Examinationkimbomd100% (2)
- SRV SLB222 en - 05062020Document2 pagesSRV SLB222 en - 05062020Nguyen ThuongNo ratings yet
- Assistive TechnologyDocument3 pagesAssistive Technologyapi-547693573No ratings yet
- Kes MahkamahDocument16 pagesKes Mahkamahfirdaus azinunNo ratings yet
- Consent CertificateDocument5 pagesConsent Certificatedhanu2399No ratings yet
- Api 579-2 - 4.4Document22 pagesApi 579-2 - 4.4Robiansah Tri AchbarNo ratings yet
- Weekly Meal Prep GuideDocument7 pagesWeekly Meal Prep Guideandrew.johnson3112No ratings yet
- Optical Fiber Communication Unit 3 NotesDocument33 pagesOptical Fiber Communication Unit 3 NotesEr SarbeshNo ratings yet
- Module 2: Environmental Science: EcosystemDocument8 pagesModule 2: Environmental Science: EcosystemHanna Dia MalateNo ratings yet
- ISBAR For Clear CommunicationDocument6 pagesISBAR For Clear Communicationmehara1920No ratings yet
- Waste Sector ProjectsDocument5 pagesWaste Sector ProjectsMrcoke SeieNo ratings yet
- Grain Silo Storage SizesDocument8 pagesGrain Silo Storage SizesTyler HallNo ratings yet
- Cooling Tower (Genius)Document7 pagesCooling Tower (Genius)JeghiNo ratings yet
- Erich FrommDocument2 pagesErich FrommTina NavarroNo ratings yet
- Pengaruh Penambahan Lateks Pada Campuran Asphalt Concrete Binder Course (AC-BC)Document10 pagesPengaruh Penambahan Lateks Pada Campuran Asphalt Concrete Binder Course (AC-BC)Haris FirdausNo ratings yet
- RestraintsDocument48 pagesRestraintsLeena Pravil100% (1)
- Hippocrates OathDocument6 pagesHippocrates OathSundary FlhorenzaNo ratings yet
- Qi Gong & Meditation - Shaolin Temple UKDocument5 pagesQi Gong & Meditation - Shaolin Temple UKBhuvnesh TenguriaNo ratings yet
- Transmission Line Loading Sag CalculatioDocument25 pagesTransmission Line Loading Sag Calculatiooaktree2010No ratings yet
- TS802 - Support StandardDocument68 pagesTS802 - Support StandardCassy AbulenciaNo ratings yet
- SSN Melaka SMK Seri Kota 2021 Annual Training Plan: Athletes Name Training ObjectivesDocument2 pagesSSN Melaka SMK Seri Kota 2021 Annual Training Plan: Athletes Name Training Objectivessiapa kahNo ratings yet
- Test Questions For Oncologic DisordersDocument6 pagesTest Questions For Oncologic Disorderspatzie100% (1)
- Gintex DSDocument1 pageGintex DSRaihanulKabirNo ratings yet
- #1 - The World'S Oldest First GraderDocument6 pages#1 - The World'S Oldest First GraderTran Van ThanhNo ratings yet
- Capacitor BanksDocument49 pagesCapacitor BanksAmal P RaviNo ratings yet